
Your Source for Cutting-Edge Research
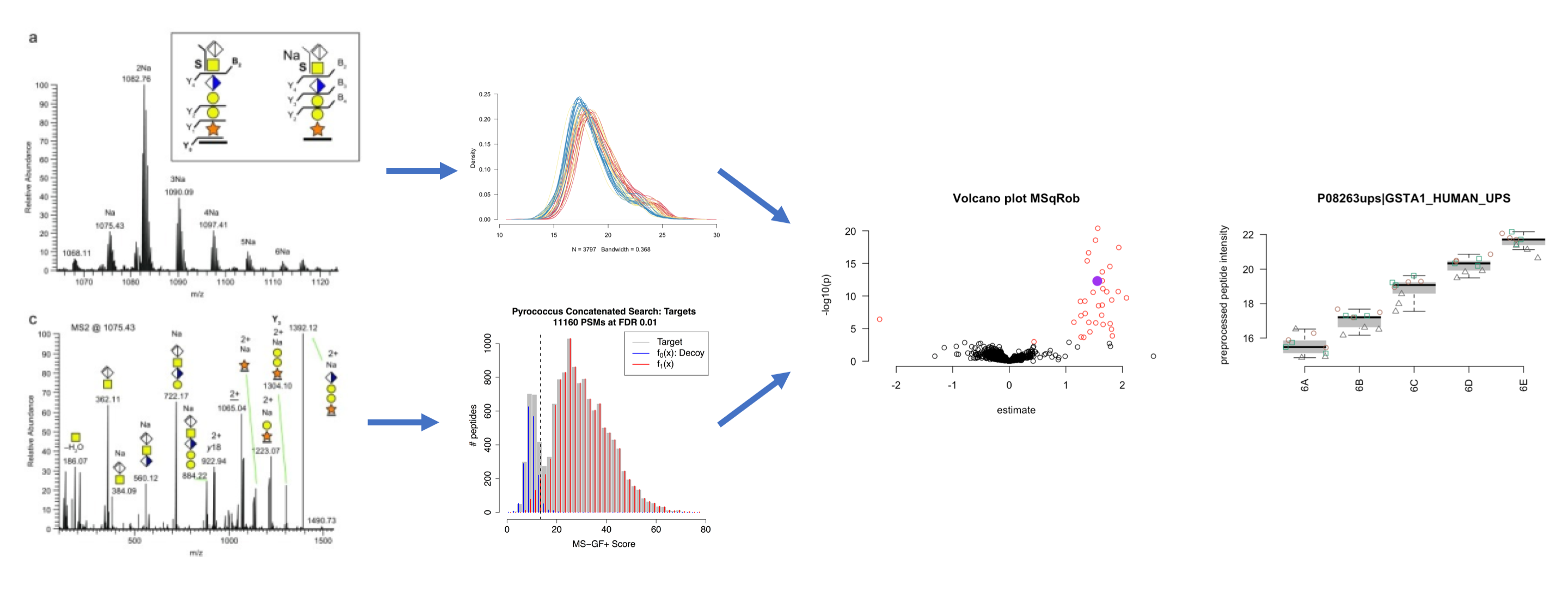
Bioinformatic analysis of proteomes starts with identifying proteins and quantifying their abundances — absolute or relative, depending on the experiment.
As with the analysis of any gene expression data, the next step is an exploratory analysis to study the variance and grouping of the data set with principal component analysis (PCA) or similar dimensionality reduction approaches.
More focused analyses may consist of differential expression and pathway analyses to characterize differences between samples.
These analyses can also be performed on proteins enriched for specific post-translational modifications such as phosphorylation. Quantitative data from the total proteome and enriched subset (e.g., phosphoproteome) can be processed in parallel or in an integrative manner to gain a more detailed view of pathway activities.
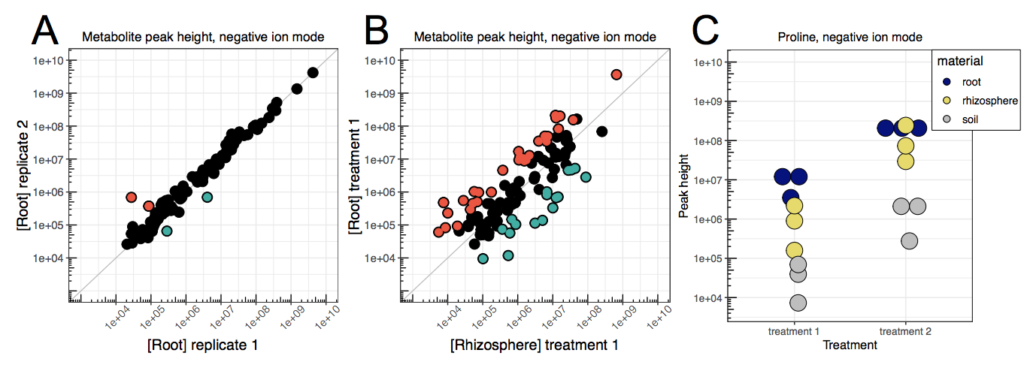
The metabolome consists of an almost endless catalog of endogenous and exogenous small molecules that partake in reactions in an organism.
While proteomics studies the catalysts of these reactions, metabolomics is concerned with their substrates, intermediates, and products.
As with proteomics, high-throughput metabolomic data is typically used to either quantify and study metabolic pathways or to identify clinically relevant molecules such as biomarkers. Thus, the explorative and statistical analyses are very similar to those of proteomics.
A special case of metabolomics, lipidomics focuses on the vast diversity of lipid molecules in an organism. Lipidomic analyses often aim at characterizing the (dys)function of lipid metabolism and trafficking, particularly in metabolic diseases.