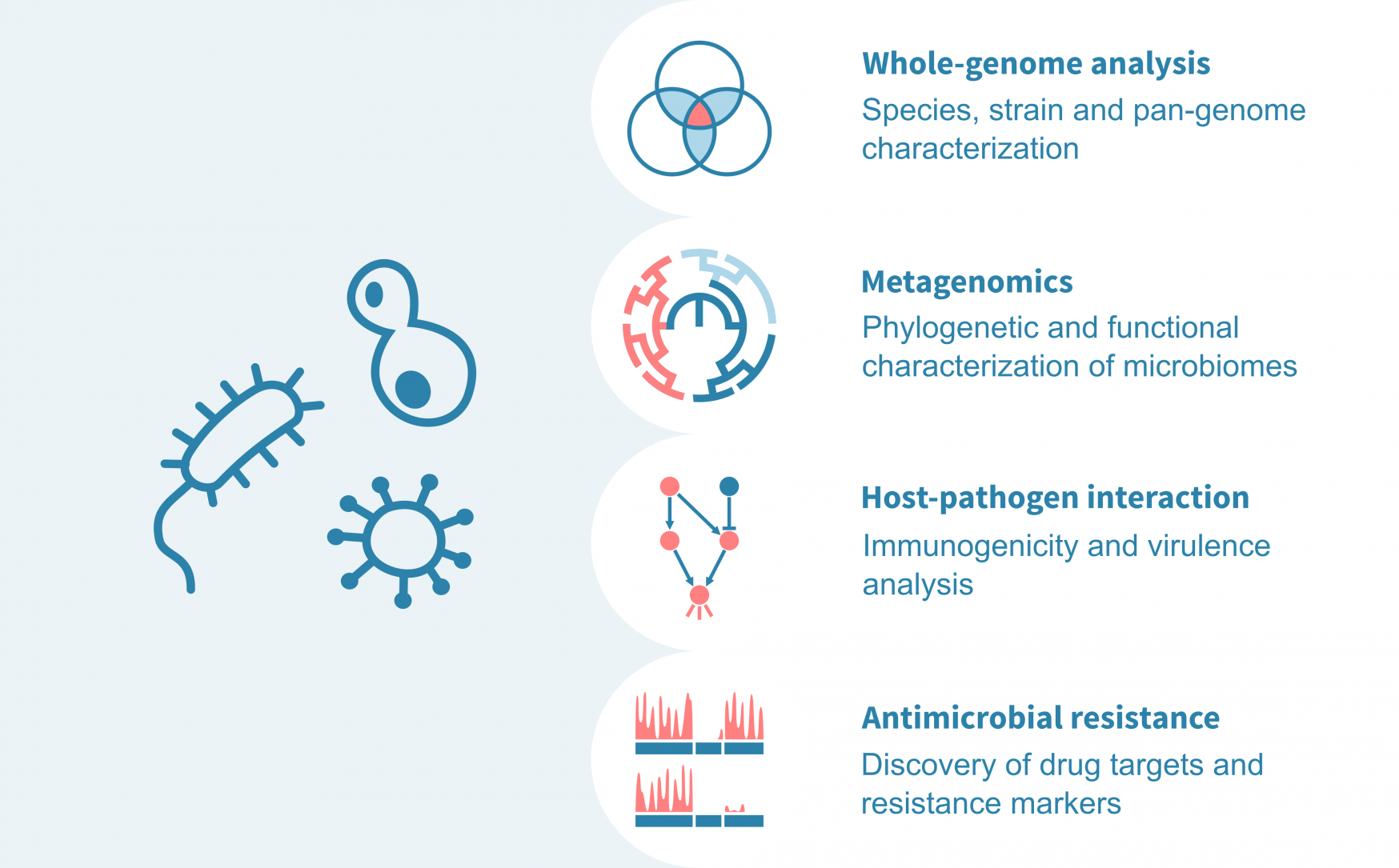
Explore microbial functions, communities and interactions.
Next-generation sequencing enables increasingly accurate identification of microbial identities, functions, and interactions.
We leverage the latest computational methods and databases to help microbiologists answer difficult questions.
How does the genomic composition of microbes affect their functions and cross-species interactions?
How can that information be used to discover new treatments or industrial applications?